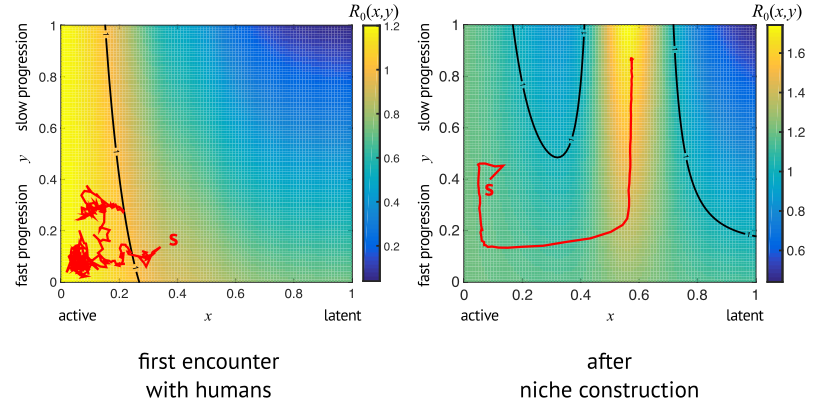
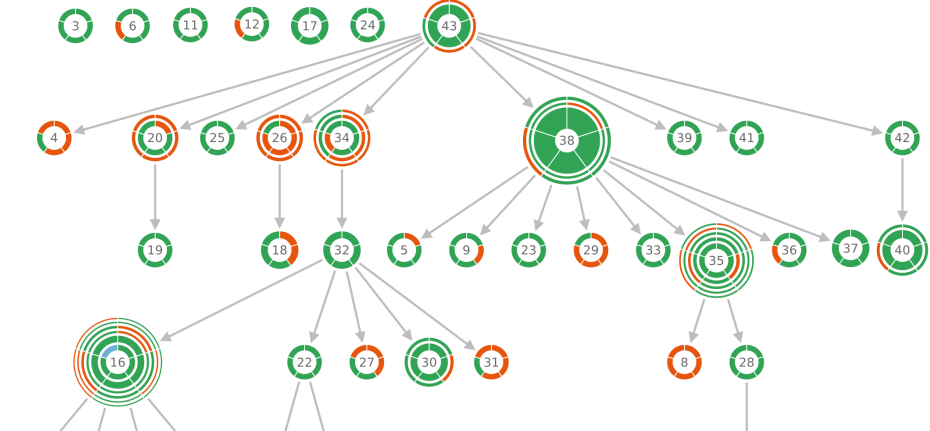
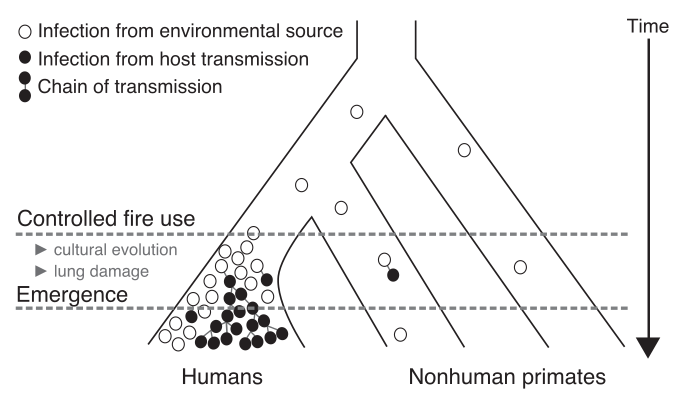
Past members
Postdocs
Sara Loo, LoÏc Thibaut, Sangeeta Bhatia, Rebecca Chisholm, Lloyd Sanders, Steven Hamblin, Frank Valckenborgh, Fabio Luciani
PhD
Xiyan Xiong, Winton Wu (primary supervisor: Jai Tree), Eden Zhang (primary supervisor: Belinda Ferrari), Yue Wu, Carmen Chan, Kayla Peck (visiting student from UNC, USA through International Practicum program), Zach Aandahl (primary supervisor: Scott Sisson), Josephine Reyes
Masters
Renault Phong, Lamia Zaghloul (visiting student from Morocco and France through International Practicum program)
Honours
Chelsea Liang, Eric Urng, Yiyi Lin, Anna Ong, Leo Fang, Justin Nam, Natalia Vaudagnotto, Alex Wong, Sara Ballouz, Carmen Chan, Alison McLean, Chaka Tang, Hui Yee Chin, Shamin Kinathil, Howard Hsu, Renault Phong, Todd Price
Undergraduate research students (internships, summer scholarships, project-based courses)
Thomas Smallbone, Maxwell Ding, Aidan McMahon-Smith, Anna Ong, Wunna Kyaw, Ian Powell, Gordon Qian, Fiona Shen, Manan Shah, James Krycer, Shamin Kinathil, AJ Joshi, Jenna Iwasenko, Hui Yee Chin, Emily Bek, Lisa Beeren
Others
Clare Saddler (visiting scientist), Yobin Jeong (Independent Learning Project program), Cuong Tran (research assistant)